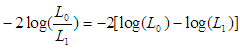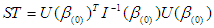Consider a clinical data set. You can download it from Here. The Diabetes data set consists nine variables. The variable Class is the response variable with a value of tested_positive and a value of tested_negative. The other eight variables are the explanatory variables.
-
#read into data
-
diabetes<-read.csv(“d:/diabetes.csv”,head=TRUE)
-
#get explanatory variables
-
#get response variable
-
y<-diabetes[2:769,9]
-
#set explantory variable’s name
- The logistic regression R code is as follows.
-
g_fun<-x%*%Beta
-
}
-
pi_value<- pi_fun(x,Beta)
-
}
-
Index<-0;
-
k<-1
-
x1<-Beta;obj<-fun(x,y,Beta);
-
objTemp<-fun(x,y,Beta)
-
Beta<-x1
-
“Results shown are based on the last maximum likelihood iteration. Validity of the model fit is questionable.”));
-
}
-
index<-1;break
-
}
-
k<-k+1
-
}
-
obj<-fun(x,y,Beta);
-
}
-
x[i]<-1e–300
-
}
-
x
-
}
-
#print(“—————–“)
-
#print(LOGL)
-
}
-
pi_value<- pi_fun(x,Beta)
-
##compute Likelihood ratio
-
MF<-ModelFitStat(x,y,Beta)
-
LR_p_value<-pchisq(LR,df,lower.tail=FALSE)
-
##compute Score
-
obj<-funs(x,y,BetaIntercept)
-
Score_p_value<-pchisq(Score,df,lower.tail=FALSE)
-
##compute Wald test
-
obj<-funs(x,y,Beta)
-
Wald_p_value<-pchisq(Wald,df,lower.tail=FALSE)
-
}
-
a<-NULL
-
}
-
}
-
a
-
}
-
}
-
}
-
}
-
}
-
i
-
}
-
NegativeFlg<-NegativeCheck(x)
-
}
-
}
-
##stepwise
-
##as matrix
-
##indication of variable
-
##intercept entered
-
indict[1]<-1
-
Beta<-NULL
-
Result<-Newtons(funs,x[,1],y)
-
Beta<-Result$Beta
-
BetaIntercept<-Result$Beta
-
indexVector<-WhichEqual1(indict)
-
##check other variable
-
VariableFlg<-NULL
-
Terminate<-FALSE
-
print(“Model building terminates because the last effect entered is removed by the Wald statistic criterion. “)
-
}
-
k<-CheckOut(k,indexVector)
-
Score_pvalue<-pchisq(Score,1,lower.tail=FALSE)
-
pvalue[i]<-Score_pvalue
-
}
-
#print(“Score pvalue for variable enter”)
-
#print(pvalue)
-
##set indication of variable
-
indict[j]<-1
-
indexVector<-WhichEqual1(indict)
-
Result<-Newtons(funs,x[,indexVector],y)
-
Beta<-NULL
-
Beta<-Result$Beta
-
##compute model fit statistics
-
MFStat<-ModelFitStat(x[,indexVector],y,Beta)
-
##test globel null hypothesis:Beta=0
-
GNTest<-GlobalNullTest(x[,indexVector],y,Beta,BetaIntercept)
-
##compute Wald test in order to remove variable
-
indexVector<-WhichEqual1(indict)
-
obj<-funs(x[,indexVector],y,Beta)
-
WaldChisqPvalue<-pchisq(WaldChisq,1,lower.tail=FALSE)
-
##check wald to decide to which variable to be removed
-
##set indication of variable
-
indict[indexVector[n+1]]<-0
-
m<- indexVector[n+1]
-
##renew Beta
-
indexVector<-WhichEqual1(indict)
-
Result<-Newtons(funs,x[,indexVector],y)
-
Beta<-NULL
-
Beta<-Result$Beta
-
Terminate<-TRUE
-
}
-
}
-
else {
-
}
-
}##repeat end
-
}
-
else {
-
}
-
}##repeat end
-
##
-
obj<-funs(x[,indexVector],y,Beta)
-
WaldChisqPvalue<-pchisq(WaldChisq,1,lower.tail=FALSE)
-
}
Finally, revoke the stepwise function.
-
Stepwise(x,y)

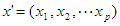 . The logic regression model is
. The logic regression model is ,
, is the probability of
is the probability of  and
and  . The right-hand side is usually referred to logic function, which can be pictured as follow.
. The right-hand side is usually referred to logic function, which can be pictured as follow.
 ,
, ,in other words, the probability of an event that not occurs.
,in other words, the probability of an event that not occurs.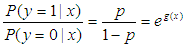 .
. .
.  . It can be achieved by maximum likelihood estimation. Since the probability of an event that occurs is
. It can be achieved by maximum likelihood estimation. Since the probability of an event that occurs is  , the probability of
, the probability of  that occurs can be writhed as
that occurs can be writhed as 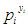 . Likewise, the probability of
. Likewise, the probability of  that occurs can be writhed as
that occurs can be writhed as  . Thus ,we can get a likelihood function
. Thus ,we can get a likelihood function
 with
with  , it can be rewritten as
, it can be rewritten as

 , we set the first order partial derivatives of
, we set the first order partial derivatives of  to zero.
to zero.

 ,for j=1,2,…p. we can use the Newton-Raphson iteration method . The matrix of second derivatives, called the Hessian, is
,for j=1,2,…p. we can use the Newton-Raphson iteration method . The matrix of second derivatives, called the Hessian, is

 denotes Hessian matrix, and
denotes Hessian matrix, and

 is an n by n diagonal matrix with element
is an n by n diagonal matrix with element 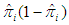 . We can get
. We can get


 directly. Since
directly. Since 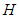 is a positive definite matrices, we can get it by Cholesky decomposition on
is a positive definite matrices, we can get it by Cholesky decomposition on  .
. is denoted as
is denoted as 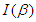 ,this matrix is called the observed information matrix. The variances and covariances of the estimated coefficients can be obtained from the inverse of
,this matrix is called the observed information matrix. The variances and covariances of the estimated coefficients can be obtained from the inverse of  , which represent as
, which represent as  .that is to say, the variance of
.that is to say, the variance of  is the
is the  diagonal element of matrix
diagonal element of matrix  , and the covariance of
, and the covariance of  and
and  , which can be denoted by
, which can be denoted by  , is the off-diagonal element. So, we can the estimated standard errors of the estimated coefficients as
, is the off-diagonal element. So, we can the estimated standard errors of the estimated coefficients as , for j=0,1,2,…,p
, for j=0,1,2,…,p 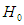 :
: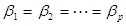 =0. We can calculate
=0. We can calculate
 is not included in
is not included in 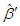 and the relevant row and column is not included in
and the relevant row and column is not included in  . This value is distributed as chi-square with
. This value is distributed as chi-square with  degrees of freedom.
degrees of freedom.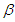 (L1) over the maximized value of the likelihood function when
(L1) over the maximized value of the likelihood function when 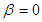 (L0). The test statistic,
(L0). The test statistic,